DNA Nanotags: A Novel Concept for 3D Model Reconstruction in Electron Microscopy
Advancing Cryo-Electron Microscopy Through Innovative Labeling
Cryo-electron microscopy has become an essential tool for protein structure analysis as resolution capabilities have improved in recent years.
The research team led by Professor Yang Yuhe at the National Center for Nanoscience focuses on understanding antigen-antibody interaction mechanisms through structural biology. Their work aims to decode the patterns of immune system responses to guide new vaccine designs and antiviral drug development.
To achieve this, the team uses cryo-electron microscopy to create three-dimensional reconstructions of various viral antigen-antibody complexes.
Challenges in Current Structural Analysis Methods
Creating high-resolution 3D models requires researchers to spend hours collecting sample images and selecting nearly a million protein particles for averaging to reduce noise and improve resolution.
A significant challenge is the difficulty in differentiating individual particles, especially when different viral subtypes have very similar morphologies. Consequently, only one specific type of complex can be loaded onto a single electron microscope grid at a time.
For viruses with numerous variants and the vast array of antibodies produced by the immune system, analyzing each complex individually demands substantial time and human resources.
The DNA Nanotag Concept: A Breakthrough Solution
To address these limitations, Professor Yang’s team introduced the concept of DNA nanotags. These programmable DNA nanostructures serve as markers with distinctive shapes that can identify each type of complex particle. This innovation allows data processing algorithms to automatically classify protein particles based on the shape of the DNA nanotags.
This multiplexed imaging approach significantly increases electron microscopy throughput, accelerating the screening and characterization of viral antigen-antibody complexes.
Applications in Cryo-Electron Tomography
Cryo-electron tomography is an emerging field in cryo-electron microscopy. This technology captures a series of images at different tilt angles of a specific area to reconstruct a 3D model of the target.
Unlike traditional cryo-electron microscopy, cryo-tomography can characterize structures in situ within cells, better reflecting the true state of protein complexes in their cellular environment.
However, this technique faces additional challenges due to lower signal-to-noise ratios (caused by the need for continuous photography with limited electron dosage per image) and the complex cellular environment that makes target particle identification and localization more difficult.
The research team believes that DNA nanotags with high contrast, substantial size, and highly specific shapes could precisely indicate target particle positions in situ, becoming a valuable tool for analyzing protein particles in their native cellular environments.
Recent Research Findings and Future Directions
The team’s recent paper, “DNA Nanotags for Multiplexed Single-Particle Electron Microscopy and In Situ Electron Cryotomography,” was published in JACS Au. Doctoral students Chen Yuanfang and Huang Yiqian are the first authors, with Professor Yang Yuhe as the corresponding author.
The paper discusses three key advantages of DNA nanostructures as nanotags:
- DNA nanostructures are highly programmable, allowing precise design of nanoscale structures in any shape using existing technologies.
- Numerous studies have demonstrated that DNA nanostructures can be easily modified with different proteins and functional molecules.
- DNA nanostructures have proven stability, maintaining structural integrity at least during the timeframe of cryo-sample preparation.
The researchers acknowledge that DNA nanotags might interfere with protein particle signals in multiplexed electron microscopy. They propose potential methods to eliminate this interference, including designing greater separation between DNA and proteins or using algorithms to weaken the DNA structure signal.
For in situ imaging, the major challenge is how DNA nanostructures can enter cells, migrate within them, and reach target sites. The paper explores possible implementation methods, including utilizing cell transmembrane transport channels, direct transmembrane delivery, and in situ expression of RNA nanostructures.
Overcoming Practical Challenges and Future Applications
According to Professor Yang, the implementation of this concept requires a “functionality first, optimization second” strategy. Initial efforts will focus on integrating existing technologies to ensure basic functionality before refining efficiency and enhancing applicability.
The team plans to design different DNA nanostructures to label different antigen subtypes of the same virus, observe them with electron microscopy after binding with antibodies, and verify whether this strategy can effectively identify protein particle types and reconstruct correct antigen-antibody binding information.
The ultimate goal is to develop DNA nanotag technology as a tool for exploring protein structures in their native state within cells, enhancing our understanding of protein function mechanisms and structure-effect relationships, which could guide nature modification and artificial manufacturing.# DNA Nanotags: A Novel Concept for 3D Model Reconstruction in Electron Microscopy
Advancing Cryo-Electron Microscopy Through Innovative Labeling
Cryo-electron microscopy has become an essential tool for protein structure analysis as resolution capabilities have improved in recent years.
The research team led by Professor Yang Yuhe at the National Center for Nanoscience focuses on understanding antigen-antibody interaction mechanisms through structural biology. Their work aims to decode the patterns of immune system responses to guide new vaccine designs and antiviral drug development.
To achieve this, the team uses cryo-electron microscopy to create three-dimensional reconstructions of various viral antigen-antibody complexes.
Challenges in Current Structural Analysis Methods
Creating high-resolution 3D models requires researchers to spend hours collecting sample images and selecting nearly a million protein particles for averaging to reduce noise and improve resolution.
A significant challenge is the difficulty in differentiating individual particles, especially when different viral subtypes have very similar morphologies. Consequently, only one specific type of complex can be loaded onto a single electron microscope grid at a time.
For viruses with numerous variants and the vast array of antibodies produced by the immune system, analyzing each complex individually demands substantial time and human resources.
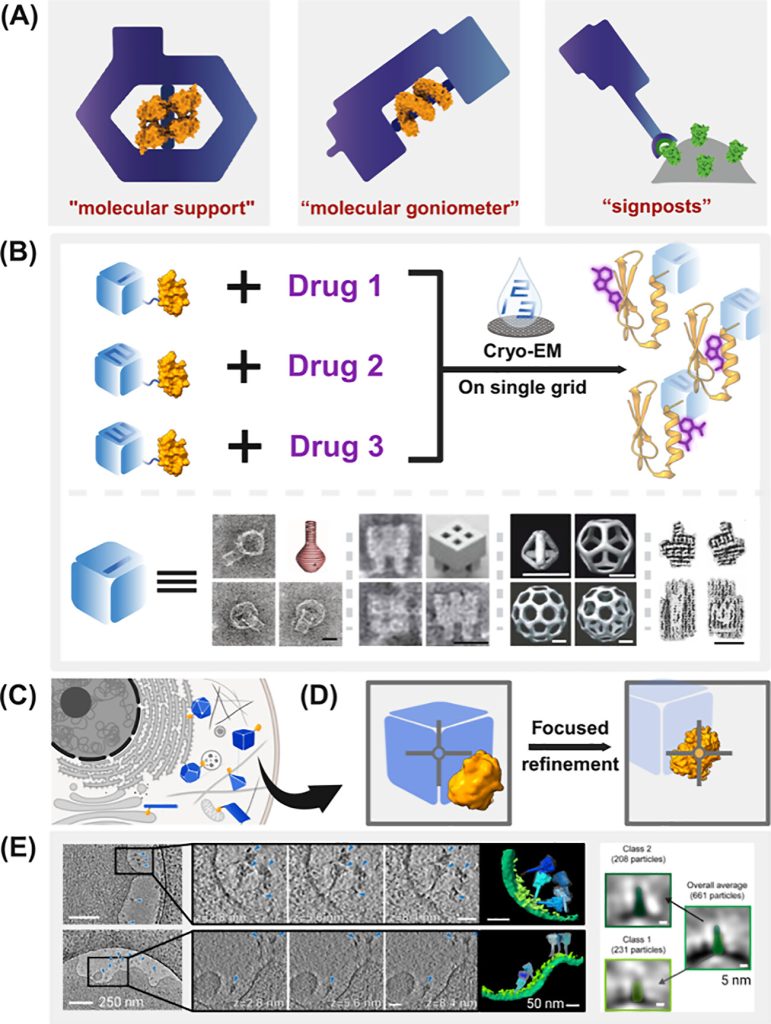
The DNA Nanotag Concept: A Breakthrough Solution
To address these limitations, Professor Yang’s team introduced the concept of DNA nanotags. These programmable DNA nanostructures serve as markers with distinctive shapes that can identify each type of complex particle. This innovation allows data processing algorithms to automatically classify protein particles based on the shape of the DNA nanotags.
This multiplexed imaging approach significantly increases electron microscopy throughput, accelerating the screening and characterization of viral antigen-antibody complexes.
Applications in Cryo-Electron Tomography
Cryo-electron tomography is an emerging field in cryo-electron microscopy. This technology captures a series of images at different tilt angles of a specific area to reconstruct a 3D model of the target.
Unlike traditional cryo-electron microscopy, cryo-tomography can characterize structures in situ within cells, better reflecting the true state of protein complexes in their cellular environment.
However, this technique faces additional challenges due to lower signal-to-noise ratios (caused by the need for continuous photography with limited electron dosage per image) and the complex cellular environment that makes target particle identification and localization more difficult.
The research team believes that DNA nanotags with high contrast, substantial size, and highly specific shapes could precisely indicate target particle positions in situ, becoming a valuable tool for analyzing protein particles in their native cellular environments.
Recent Research Findings and Future Directions
The team’s recent paper, “DNA Nanotags for Multiplexed Single-Particle Electron Microscopy and In Situ Electron Cryotomography,” was published in JACS Au. Doctoral students Chen Yuanfang and Huang Yiqian are the first authors, with Professor Yang Yuhe as the corresponding author.
The paper discusses three key advantages of DNA nanostructures as nanotags:
- DNA nanostructures are highly programmable, allowing precise design of nanoscale structures in any shape using existing technologies.
- Numerous studies have demonstrated that DNA nanostructures can be easily modified with different proteins and functional molecules.
- DNA nanostructures have proven stability, maintaining structural integrity at least during the timeframe of cryo-sample preparation.
The researchers acknowledge that DNA nanotags might interfere with protein particle signals in multiplexed electron microscopy. They propose potential methods to eliminate this interference, including designing greater separation between DNA and proteins or using algorithms to weaken the DNA structure signal.
For in situ imaging, the major challenge is how DNA nanostructures can enter cells, migrate within them, and reach target sites. The paper explores possible implementation methods, including utilizing cell transmembrane transport channels, direct transmembrane delivery, and in situ expression of RNA nanostructures.
Overcoming Practical Challenges and Future Applications
According to Professor Yang, the implementation of this concept requires a “functionality first, optimization second” strategy. Initial efforts will focus on integrating existing technologies to ensure basic functionality before refining efficiency and enhancing applicability.
The team plans to design different DNA nanostructures to label different antigen subtypes of the same virus, observe them with electron microscopy after binding with antibodies, and verify whether this strategy can effectively identify protein particle types and reconstruct correct antigen-antibody binding information.
The ultimate goal is to develop DNA nanotag technology as a tool for exploring protein structures in their native state within cells, enhancing our understanding of protein function mechanisms and structure-effect relationships, which could guide nature modification and artificial manufacturing.